检测到您当前使用浏览器版本过于老旧,会导致无法正常浏览网站;请您使用电脑里的其他浏览器如:360、QQ、搜狗浏览器的极速模式浏览,或者使用谷歌、火狐等浏览器。
 下载Firefox
下载Firefox
检测到您当前使用浏览器版本过于老旧,会导致无法正常浏览网站;请您使用电脑里的其他浏览器如:360、QQ、搜狗浏览器的极速模式浏览,或者使用谷歌、火狐等浏览器。
 下载Firefox
下载Firefox
作为哺乳动物体内酸性最强的器官,胃所含的强酸性胃液(pH值为1-3)是人和动物抵御绝大多数微生物病菌的一道天然屏障。然而,肠道病原菌能够在强酸性的胃液下存活,并进而造成肠道感染。我院昌增益课题组化学院陈鹏课题组(二者都属于北大跨院系的蛋白质科学中心成员)通过合作研究,系统地捕获了一种酸性分子伴侣蛋白在酸胁迫下的“客户蛋白”,并依此阐释了大肠杆菌抵御胃酸的机理。 这一结果于2011年9月4日以Article的形式,在自然杂志子刊“Nature Chemical Biology”发表:
Zhang M, et al “A genetically incorporated crosslinker reveals chaperone cooperation in acid resistance” doi:10.1038/nchembio.644 (第一作者为昌增益教授的前博士生张萌)
http://www.nature.com/nchembio/index.html
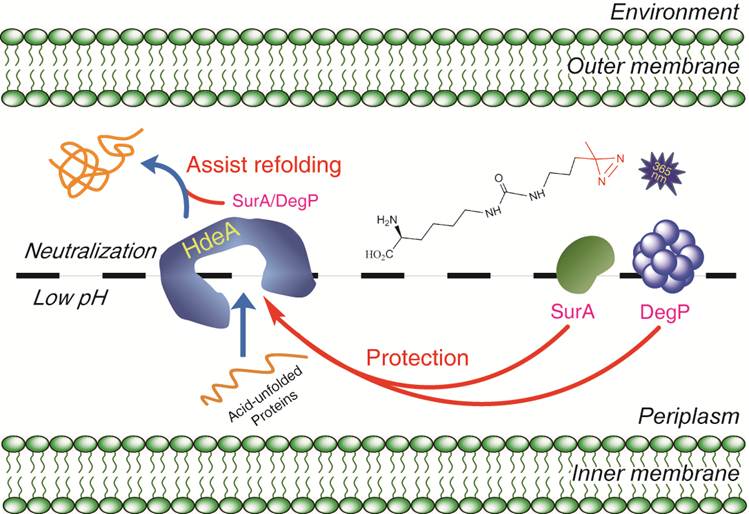
大肠杆菌的变异菌株依旧严重地威胁着人类的健康,尤其是最近源于德国的致死性H104:04型大肠杆菌的爆发,在整个欧洲都造成了恐慌。对于包括大肠杆菌和志贺氏菌在内的肠 道病原体微生物来说,顺利地通过人体胃液是它们对肠道进行感染的先决条件。因此,深刻理解大肠杆菌的抗酸性机理将极大地加深我们对这类病原菌的认识,为今 后发展新型抗生素奠定基础。通过开发一种全新的蛋白质光交联探针,他们成功地捕捉了大肠杆菌体内的一个关键酸性分子伴侣HdeA的作用底物。对其中的两个本身也是分子伴侣的关键“客户蛋白”DegP 和SurA的进一步研究,使他们发现了一种不依赖于ATP的“分子伴侣-保护分子伴侣”的独特机制,并证明细菌利用这一作用来增加其逃逸胃酸防线的成功率。该文章所描述的一种不以ATP作为能量货币的分子伴侣间相互作用,以及细菌通过这一相互作用来抵抗胃酸的全新发现都在国际上属首次报道,一经发表就引起了广泛的关注。 Nature Asia-Pacific在2011年9月5日在其网站上作为亮点介绍了该工作,美国化学会的官方杂志《化学与化工新闻》(Chemical & Engineering News)以News Coverage的形式于2011年9月7日报道了这一工作(相关内容见下)。
此项研究是在昌增益课题组之前的本科生洪玮哲和吴叶(两人现都在美国斯坦福大学攻读博士学位)相继开展的研究基础上的继续。洪玮哲之前的工作发现,肠道细菌抗酸分子伴侣蛋白质HdeA发挥活性时,利用的是一种三维空间结构大部分丧失的独特机制(Hong Weizhe et al.“Periplasmic Protein HdeA Exhibits Chaperone-like Activity Exclusively within Stomach pH Range by Transforming into Disordered Conformation”, J. Biol. Chem., 2005, 280:27029-27034)。吴叶后来的工作表明,蛋白质处于伸展状态时的肽链双亲性(分子的部分亲水和另一部分亲脂)结构特征对该蛋白质赋予肠道细菌抗酸功能是必须的(Wu Ye et al., “Conserved Amphiphilic Feature Is Essential for Periplasmic Chaperone HdeA to Support Acid Resistance in Enteric Bacteria”, Biochem. J., 2008, 412:389-397).
陈鹏课题组在之前的工作中系统地阐述了利用非天然氨基酸进行蛋白质特异标记和研究蛋白质相互作用的手段(Hao Ziyang et al, Acc. Chem. Res. “Introducing Bioorthogonal Functionalities into Proteins in Living Cells”, Acc. Chem. Res., 2011 DOI: 10.1021/ar200067r)。这次报道的光交联探针能够广泛地用于捕捉活细胞体内的蛋白质-蛋白质相互作用。
该研究得到了科技部、国家自然科学基金委和英国威廉希尔公司985计划的资助。

Nature Chemical Biology, September 5, 2011
http://www.natureasia.com/en/highlights/details.php?id=1408
A cooperative group of proteins that work together to protect cells from acid stress has been found in bacteria that transit through the human digestive tract, as reported online this week in Nature Chemical Biology.
When proteins encounter highly acidic environments, their normally stable structure can fall apart; if enough proteins lose their structure, the cell cannot operate normally and will die. Chaperones are a family of proteins that protect other proteins from losing their structure or help the proteins to regain their structure once lost.
Zengyi Chang, Peng Chen and colleagues look for ‘clients’ – or preferred protein substrates – of one of these chaperones by using a newly-designed non-natural amino acid to link the chaperone and its client together, allowing them to be identified as interacting. Among the clients, the authors discovered two more chaperones that subsequent tests demonstrated were working together with the initial chaperone to help other proteins more quickly regain their shape. This cooperative mechanism explains how bacteria might recover from their acidic travels.

September 7, 2011
Bacterial Acid Trips
Chemical Biology: New technique reveals how pathogens endure our acidic stomachs
http://pubs.acs.org/cen/news/89/i37/8937notw6.html
|
|
The side chain of this artificial amino acid possesses an alkyl diazirine group that can crosslink to proteins in acidic conditions when exposed to light.
|
When you get food poisoning, the bacteria causing havoc in your intestines have first navigated a rather treacherous journey through your acidic stomach. The molecular mechanisms by which a pathogen survives this acid environment have long kept researchers guessing, but now a team of biochemists led by Peng R. Chen at Peking University in China have developed a new technique to study the bacterial coping mechanism (Nature Chem. Biol., DOI: 10.1038/NChemBio.644).
In addition to helping researchers better understand pathogen survival, the new technique, which permits protein-protein interactions to be studied at low pH, will likely find application in probing a wide range of biology that occurs in acidic conditions. To date, studying protein-protein interactions at low pH has been a challenge because most current techniques don`t work in strong acidic environments, Chen explains.
To study how Escherichia coli survives our stomach, Chen`s group, in collaboration with Zengyi Chang, also at Peking University, focused on the bacteria`s chaperone proteins, which keep cellular proteins from unfolding in harmful environments. They engineered the gene for an essential chaperone protein called HdeA to incorporate an artificial amino acid at the site that binds client proteins. The artificial amino acid has a side chain that possesses an alkyl diazirine moiety that can crosslink to other proteins when irradiated.
The team exposed this engineered E. coli to acid so HdeA would start protecting proteins from denaturation. Then they hit the bacterium with light to trap HdeA with its client proteins and used mass spectrometry to identify these clients.
Among the dozens of client proteins protected by HdeA, the team detected two other chaperone proteins. This suggests that HdeA is a mother chaperone protein in a large acid-resistance network, note the authors. They also found that HdeA could protect E. coli cells without the help of adenosine triphosphate, the common chemical energy currency in cells.
It`s "a very clever and elegant method," says John Foster, a microbiologist who studies pathogen acid resistance at the University of South Alabama. "The real value of this work is in the methodology, which could be used to probe many other chaperone-client interactions in bacteria, archaea, and eukaryotes."
Indeed, Chen says he`s currently using the approach to examine protein-protein interactions in the lysosome, the organelle in eukaryotes whose acidic interior is responsible for breaking down waste proteins and other cellular molecules.